My Potential Comorbid ADHD-Dyslexia Risk Factors based on 2023 Published 'Associative gene networks reveal novel candidates important for ADHD and dyslexia comorbidity'
I read a Genome Wide Association Study about associative gene networks revealing novel candidates important for ADHD and dyslexia comorbidity.
I was very interested because of my being being a neurodivergent with Dyslexia, Dyspraxia, ADHD as well as Ataxia which is a rare neurological condition that involve problems with coordination like Dyspraxia does.
Abstract
Background
Attention deficit hyperactivity disorder (ADHD) is commonly associated with developmental dyslexia (DD), which are both prevalent and complicated pediatric neurodevelopmental disorders that have a significant influence on children’s learning and development. Clinically, the comorbidity incidence of DD and ADHD is between 25 and 48%. Children with DD and ADHD may have more severe cognitive deficiencies, a poorer level of schooling, and a higher risk of social and emotional management disorders. Furthermore, patients with this comorbidity are frequently treated for a single condition in clinical settings, and the therapeutic outcome is poor. The development of effective treatment approaches against these diseases is complicated by their comorbidity features. This is often a major problem in diagnosis and treatment. In this study, we developed bioinformatical methodology for the analysis of the comorbidity of these two diseases. As such, the search for candidate genes related to the comorbid conditions of ADHD and DD can help in elucidating the molecular mechanisms underlying the comorbid condition, and can also be useful for genotyping and identifying new drug targets.
Results
Using the ANDSystem tool, the reconstruction and analysis of gene networks associated with ADHD and dyslexia was carried out. The gene network of ADHD included 599 genes/proteins and 148,978 interactions, while that of dyslexia included 167 genes/proteins and 27,083 interactions. When the ANDSystem and GeneCards data were combined, a total of 213 genes/proteins for ADHD and dyslexia were found. An approach for ranking genes implicated in the comorbid condition of the two diseases was proposed. The approach is based on ten criteria for ranking genes by their importance, including relevance scores of association between disease and genes, standard methods of gene prioritization, as well as original criteria that take into account the characteristics of an associative gene network and the presence of known polymorphisms in the analyzed genes. Among the top 20 genes with the highest priority DRD2, DRD4, CNTNAP2 and GRIN2B are mentioned in the literature as directly linked with the comorbidity of ADHD and dyslexia. According to the proposed approach, the genes OPRM1, CHRNA4 and SNCA had the highest priority in the development of comorbidity of these two diseases. Additionally, it was revealed that the most relevant genes are involved in biological processes related to signal transduction, positive regulation of transcription from RNA polymerase II promoters, chemical synaptic transmission, response to drugs, ion transmembrane transport, nervous system development, cell adhesion, and neuron migration.
Conclusions
The application of methods of reconstruction and analysis of gene networks is a powerful tool for studying the molecular mechanisms of comorbid conditions. The method put forth to rank genes by their importance for the comorbid condition of ADHD and dyslexia was employed to predict genes that play key roles in the development of the comorbid condition. The results can be utilized to plan experiments for the identification of novel candidate genes and search for novel pharmacological targets.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10478365/
I looked at variants with a common allele frequency of no more than 5%.
They are based on Grpmax Filtering AF in gnomAD v4
This annotation can be used for filtering variants by allele frequency against a disease-specific threshold that can be set for each disease (e.g. BA1 in the 2015 ACMG/AMP guidelines). In this case the filtering allele frequency (FAF) is the maximum credible genetic ancestry group AF (e.g. the lower bound of the 95% confidence interval (CI)). If the FAF is above the disease-specific threshold, then the observed AC is not compatible with pathogenicity. See http://cardiodb.org/allelefrequencyapp/ and Whiffin et al. 2017 for additional information.
https://gnomad.broadinstitute.org/help/faf
I looked for variants with Combined Annotation Dependent Depletion (CADD) scores of at least 10, and so the variants are predicted to be in the top 10% of the most deleterious.
My focus was mainly on the 5 Prime Untranslated Region, 3 Prime Untranslated Region, and Promoter because they are critical regulatory features in genes. I used VEP to see if the variants involve these regulatory features.
exceptions in regards to CADD scores: 5 Prime Untranslated Region (5'UTR) perturbing variants (Premature Start Codon Gain, Premature Start Codon Loss, Premature Stop Codon Gain, Premature Stop Codon Loss, uORF Frameshift)
In molecular genetics, an untranslated region (or UTR) refers to either of two sections, one on each side of a coding sequence on a strand of mRNA. If it is found on the 5' side, it is called the 5' UTR (or leader sequence), or if it is found on the 3' side, it is called the 3' UTR (or trailer sequence). mRNA is RNA that carries information from DNA to the ribosome, the site of protein synthesis (translation) within a cell. The mRNA is initially transcribed from the corresponding DNA sequence and then translated into protein. However, several regions of the mRNA are usually not translated into protein, including the 5' and 3' UTRs.
Although they are called untranslated regions, and do not form the protein-coding region of the gene, uORFs located within the 5' UTR can be translated into peptides.[1]
The 5' UTR is upstream from the coding sequence. Within the 5' UTR is a sequence that is recognized by the ribosome which allows the ribosome to bind and initiate translation. The mechanism of translation initiation differs in prokaryotes and eukaryotes. The 3' UTR is found immediately following the translation stop codon. The 3' UTR plays a critical role in translation termination as well as post-transcriptional modification.[2]
These often long sequences were once thought to be useless or junk mRNA that has simply accumulated over evolutionary time. However, it is now known that the untranslated region of mRNA is involved in many regulatory aspects of gene expression in eukaryotic organisms. The importance of these non-coding regions is supported by evolutionary reasoning, as natural selection would have otherwise eliminated this unusable RNA.
It is important to distinguish the 5' and 3' UTRs from other non-protein-coding RNA. Within the coding sequence of pre-mRNA, there can be found sections of RNA that will not be included in the protein product. These sections of RNA are called introns. The RNA that results from RNA splicing is a sequence of exons. The reason why introns are not considered untranslated regions is that the introns are spliced out in the process of RNA splicing. The introns are not included in the mature mRNA molecule that will undergo translation and are thus considered non-protein-coding RNA.
https://en.wikipedia.org/wiki/Untranslated_region
Ensembl youtube video playlist Gene Regulation - 8 videos
https://www.youtube.com/playlist?list=PLqB8Yx1tGBMbFtUj_3rYxoasRiMqe3ltO
Top 20 genes considered in Dyslexia-ADHD comorbidity
1. OPRM1
2. CHRNA4
3. SNCA
4. GRIN1
5. MAPT
6. DRD2
7. DLG4
8. GABRA6
9. GRM3
10. ESR1
11. DRD4
12. CACNA1A - one of the genes known to be associated with Ataxia
https://www.mayocliniclabs.com/test-catalog/overview/617506
13. GABBR1
14. CNTNAP2
15. SCN1A
16. GABRA1
17. DRD5
18. IL6
19. GRIN2B
20. PICK1
GABRA6 (Gamma-Aminobutyric Acid Type A Receptor Subunit Alpha6) is one of the genes in 5q34 (chr5:160500001-169000000) which was listed as one of the 42 genome-wide significant loci associated with dyslexia. I have a GABRA6 intronic variant listed in my Dyslexia GWAS blog post.
I have a DRD4 (Dopamine Receptor D4) 5 Prime Untranslated Region (5'UTR) Promoter variant that I inherited from my mother that is reported in ClinVar as Uncertain Significance for hereditary ADHD. It has a condition allele frequency of more than 5% which is the maximum that I use for ADHD, and so I did not include the variant in my list of potential comorbid Dyslexia-ADHD risk factors. It has a condition allele frequency of less than 10% which is the maximum that I use for Dyslexia, and so it could be a potential risk factor for Dyslexia as well as an indicator of novelty seeking personality.
https://neurodivergence.blogspot.com/2024/01/the-drd4-dopamine-receptor-d4-gene.html
Considering all my potential Dyslexia and ADHD risk factor variants based on the three Genome Wide Association Studies
2022 Published GWAS Dyslexia
I have 4 TANC2 variants, 3 CALN1 variants, 1 GGNBP2 variant, 1 TMEM182 variant, 1 MITF variant, 1 SGCD variant, 1 SEMA3F variant, 1 GABRA6 variant for a total of 13 potential risk factor variants.
https://neurodivergence.blogspot.com/2024/02/my-potential-dyslexia-risk-factors.html
2022 Published GWAS ADHD
I have 1 FOXP2 variant, 1 MON1A variant 1 ELOVL1 variant, 1 RHOA variant, 1 CDH8 variant, 2 CALN1 variants for a total of 6 potential risk factor variants.
https://neurodivergence.blogspot.com/2024/03/my-potential-adhd-risk-factors-based-on.html
2023 Published GWAS ADHD-Dyslexia Comorbidity
I have 1 ESR1 variant, 1 MAPT variant, 1 DLG4 variant, 1 GABRA6 variant, 1 OPRM1 variant for a total of 5 potential risk factor variants.
I have 17 potential Dyslexia risk factor variants that can factor into polygenic nature of my Dyslexia and 12 potential ADHD risk factor variants that can factor into polygenic nature of my ADHD.
This is the snapshot of my Dante Labs account with my genome number
My Sequencing Genomic Data
My Dante Labs Genomic Data
ESR1
ESR1 (Estrogen Receptor 1) is a Protein Coding gene. Diseases associated with ESR1 include Estrogen Resistance and Breast Cancer. Among its related pathways are PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling and ESR-mediated signaling. Gene Ontology (GO) annotations related to this gene include DNA-binding transcription factor activity and identical protein binding. An important paralog of this gene is ESR2.
This gene encodes an estrogen receptor and ligand-activated transcription factor. The canonical protein contains an N-terminal ligand-independent transactivation domain, a central DNA binding domain, a hinge domain, and a C-terminal ligand-dependent transactivation domain. The protein localizes to the nucleus where it may form either a homodimer or a heterodimer with estrogen receptor 2. The protein encoded by this gene regulates the transcription of many estrogen-inducible genes that play a role in growth, metabolism, sexual development, gestation, and other reproductive functions and is expressed in many non-reproductive tissues. The receptor encoded by this gene plays a key role in breast cancer, endometrial cancer, and osteoporosis. This gene is reported to have dozens of transcript variants due to the use of alternate promoters and alternative splicing, however, the full-length nature of many of these variants remain uncertain. [provided by RefSeq, Jul 2020]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=ESR1
rs11963577 6-151804922-C-T
5'UTR Premature Start Codon Gain Variant
25 out of 152,342 (0.01641%) Condition allele frequency is 1.181%
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA150367014
MAPT
MAPT (Microtubule Associated Protein Tau) is a Protein Coding gene. Diseases associated with MAPT include Frontotemporal Dementia and Supranuclear Palsy, Progressive, 1. Among its related pathways are Transmission across Chemical Synapses and Apoptotic cleavage of cellular proteins. Gene Ontology (GO) annotations related to this gene include protein kinase binding and microtubule binding. An important paralog of this gene is MAP2.
This gene encodes the microtubule-associated protein tau (MAPT) whose transcript undergoes complex, regulated alternative splicing, giving rise to several mRNA species. MAPT transcripts are differentially expressed in the nervous system, depending on stage of neuronal maturation and neuron type. MAPT gene mutations have been associated with several neurodegenerative disorders such as Alzheimer's disease, Pick's disease, frontotemporal dementia, cortico-basal degeneration and progressive supranuclear palsy. [provided by RefSeq, Jul 2008]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=MAPT
rs10593245 17-45895048-CGTGTGTGTGT-C (10 base pair deletion)
Intronic Variant in Promoter
86 out of 133,562 (0.06439%) Condition allele frequency is 0.1631%
14.5
Mom doesn't have it
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA772438015
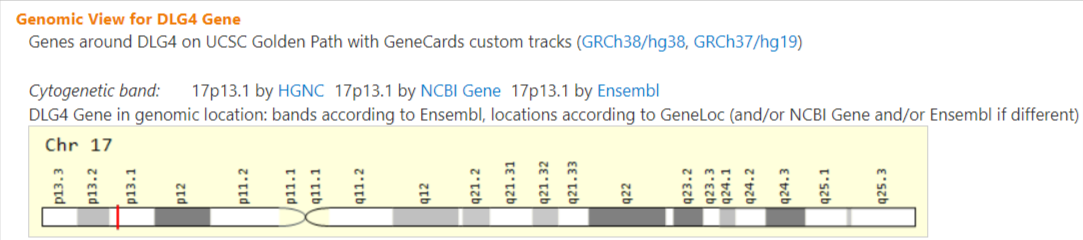
DLG4
DLG4 (Discs Large MAGUK Scaffold Protein 4) is a Protein Coding gene. Diseases associated with DLG4 include Intellectual Developmental Disorder, Autosomal Dominant 62 and Acyl-Coa Dehydrogenase, Very Long-Chain, Deficiency Of. Among its related pathways are Unblocking of NMDA receptors, glutamate binding and activation and RAF/MAP kinase cascade. Gene Ontology (GO) annotations related to this gene include signaling receptor binding and obsolete protein C-terminus binding. An important paralog of this gene is DLG1.
This gene encodes a member of the membrane-associated guanylate kinase (MAGUK) family. It heteromultimerizes with another MAGUK protein, DLG2, and is recruited into NMDA receptor and potassium channel clusters. These two MAGUK proteins may interact at postsynaptic sites to form a multimeric scaffold for the clustering of receptors, ion channels, and associated signaling proteins. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=DLG4
rs114403556 17-7194528-G-A
Intronic Variant
1,759 out of 1,575,562 (0.1116%) Condition allele frequency is 2.005%
14.1
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA8336980
GABRA6
GABRA6 (Gamma-Aminobutyric Acid Type A Receptor Subunit Alpha6) is a Protein Coding gene. Diseases associated with GABRA6 include Anxiety and Intraventricular Meningioma. Among its related pathways are Transmission across Chemical Synapses and GABA B receptor activation. Gene Ontology (GO) annotations related to this gene include chloride channel activity and GABA-A receptor activity. An important paralog of this gene is GABRA4.
GABA is the major inhibitory neurotransmitter in the mammalian brain where it acts at GABA-A receptors, which are ligand-gated chloride channels. Chloride conductance of these channels can be modulated by agents such as benzodiazepines that bind to the GABA-A receptor. At least 16 distinct subunits of GABA-A receptors have been identified. [provided by RefSeq, Jul 2008]
rs937339709 5-161700233-G-A
Intronic Variant
25 out of 152,342 (0.01641%) Condition allele frequency is 0.09184%
13.6
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA131062740
OPRM1
OPRM1 (Opioid Receptor Mu 1) is a Protein Coding gene. Diseases associated with OPRM1 include Substance Dependence and Drug Dependence. Among its related pathways are Class A/1 (Rhodopsin-like receptors) and Gene expression (Transcription). Gene Ontology (GO) annotations related to this gene include G protein-coupled receptor activity and voltage-gated calcium channel activity. An important paralog of this gene is OPRK1.
This gene encodes one of at least three opioid receptors in humans; the mu opioid receptor (MOR). The MOR is the principal target of endogenous opioid peptides and opioid analgesic agents such as beta-endorphin and enkephalins. The MOR also has an important role in dependence to other drugs of abuse, such as nicotine, cocaine, and alcohol via its modulation of the dopamine system. The NM_001008503.2:c.118A>G allele has been associated with opioid and alcohol addiction and variations in pain sensitivity but evidence for it having a causal role is conflicting. Multiple transcript variants encoding different isoforms have been found for this gene. Though the canonical MOR belongs to the superfamily of 7-transmembrane-spanning G-protein-coupled receptors some isoforms of this gene have only 6 transmembrane domains. [provided by RefSeq, Oct 2013]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=OPRM1
rs535375194 6-154098833-T-C
Intronic Variant
57 out of 152,334 (0.03742%) Condition allele frequency is 0.1065%
10.0
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA150254785







.png)
.png)


Comments