My Potential Dyslexia Risk Factors Based On 2022 Published 'Discovery of 42 genome-wide significant loci associated with dyslexia'
I read a Genome Wide Association Study about a discovery of 42 genome-wide significant loci associated with dyslexia. With my being a neurodivergent with Dyslexia, Dyspraxia, and Attention Deficit-Hyperactivity Disorder (ADHD) as well as having Ataxia, I was very interested. I was mistook for being intellectually handicapped and misplaced in a special education class with intellectually disabled children in 1st grade after being in a regular class in kindergarten. My mother even got her Fallopian Tubes tied because she felt that I would need a lot of special attention and care. I was placed in a more appropriate special education class in 2nd grade after I was found to be of average above intelligence. I repeated the 2nd grade. My mother told me that it was her decision. She thought I wasn't mature enough. She told me that I had the possiblity skip a grade. I started being mainstreamed in regular classroom around the end of 2nd grade. I was full mainstream student from 3rd grade to 12th grade. I was predicted to be mainstreamed in regular classes in 6th grade. I had auditory therapy, speech therapy, phonics training, and motor skills therapy to correct my Dyslexic and Dyspraxic weaknesses. I was diagnosed ADHD, Inattentive type in 2004. I was diagnosed as having Cerebellar Vestibular Dysfunction by world-renowned psychiatrist/neurologist Dr. Harold N. Levinson in 2005. I was examined by Veteran Affairs neurologists and neuropsychologists in 2006, and my Dyslexia and Dyspraxia were confirmed. My Veteran Affairs Problem List record lists Ataxia, Dyslexia, Abnormal Auditory Perception, Saccadic Eye Movement Deficiency, and Smooth Pursuit Movement Deficiency. Because of my speech and coordination problems, I was examined by neurologists when I was 3 years old. They told my mother that I didn't have any brain damage. My VA neurological testing revealed no brain damage. They noted my neurological problems were genetic and not acquired for I didn't have progression of symptoms. My neurological problems got milder as the result of early intervention therapies, but I am aware that my neurological problems could get worse in my old age.
I read about Ataxia. For hereditary ataxias, the prevalence rate is 10 cases per 100,000 individuals. In children, the prevalence of ataxia is 26 cases per 100,000 individuals.
https://neurodivergence.blogspot.com/2023/12/my-indicators-for-left-handedness-and.html
Abstract
Reading and writing are crucial life skills but roughly one in ten children are affected by dyslexia, which can persist into adulthood. Family studies of dyslexia suggest heritability up to 70%, yet few convincing genetic markers have been found. Here we performed a genome-wide association study of 51,800 adults self-reporting a dyslexia diagnosis and 1,087,070 controls and identified 42 independent genome-wide significant loci: 15 in genes linked to cognitive ability/educational attainment, and 27 new and potentially more specific to dyslexia. We validated 23 loci (13 new) in independent cohorts of Chinese and European ancestry. Genetic etiology of dyslexia was similar between sexes, and genetic covariance with many traits was found, including ambidexterity, but not neuroanatomical measures of language-related circuitry. Dyslexia polygenic scores explained up to 6% of variance in reading traits, and might in future contribute to earlier identification and remediation of dyslexia.
Main
The ability to read is crucial for success at school and access to employment, information and health and social services, and is related to attained socioeconomic status1. Dyslexia is a neurodevelopmental disorder characterized by severe reading difficulties, present in 5–17.5% of the population, depending on diagnostic criteria2,3. It often involves impaired phonological processing (the decoding of sound units, or phonemes, within words) and frequently co-occurs with psychiatric and other developmental disorders4, especially attention-deficit hyperactivity disorder (ADHD)5,6 and speech and language disorders7,8. Dyslexia may represent the low extreme of a continuum of reading ability, a complex multifactorial trait with heritability estimates ranging from 40% to 80%9,10. Identifying genetic risk factors not only aids increased understanding of the biological mechanisms, but may also expand diagnostic capabilities, facilitating earlier identification of individuals prone to dyslexia and co-occurring disorders for specific support.
Previous genome-wide investigations of dyslexia have been limited to linkage analyses of affected families11 or modest (n < 2,300 cases) association studies of diagnosed children and adolescents12. Candidate genes from linkage studies show inconsistent replication, and genome-wide association studies (GWAS) have not found significant associations, although LOC388780 and VEPH1 were supported in gene-based tests12. Larger cohorts are vital for increasing sensitivity to detect new genetic associations of small effect. Here, we present the largest dyslexia GWAS to date, with 51,800 adults self-reporting a dyslexia diagnosis and 1,087,070 controls, all of whom are research participants with the personal genetics company 23andMe, Inc. We validate our association discoveries in independent cohorts, provide functional annotations of significant variants (mainly single-nucleotide polymorphisms (SNPs)) and potential causal genes, and estimates of SNP-based heritability. Lastly, we investigate genetic correlations with reading and related skills, health, socioeconomic, and psychiatric measures, and evaluate the evidence for previously implicated dyslexia candidate genes in our well-powered results.
Results
Genome-wide associations
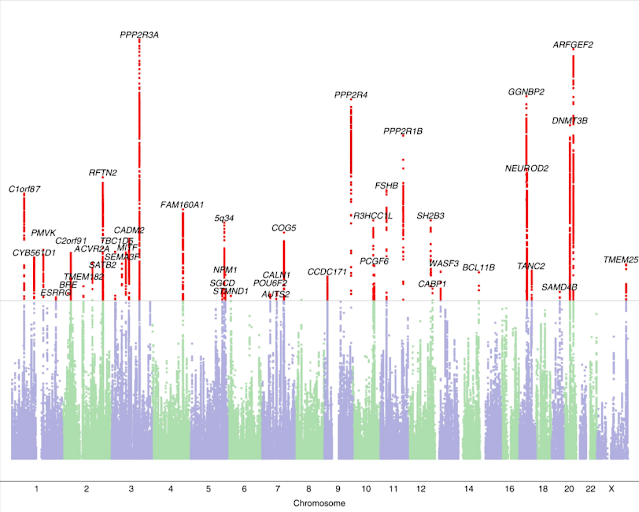
The full dataset included 51,800 (21,513 males, 30,287 females) participants responding ‘yes’ to the question ‘Have you been diagnosed with dyslexia?’ (cases) and 1,087,070 (446,054 males, 641,016 females) participants responding ‘no’ (controls). Participants were aged 18 years or over (mean ages of cases and controls were 49.6 years (s.d. 16.2) and 51.7 years (s.d. 16.6), respectively). We identified 42 independent genome-wide significant associated loci (P < 5 × 10−8) and 64 loci with suggestive significance (P < 1 × 10−6) (Fig. 1 and Supplementary Table 1). Genomic inflation was moderate (λGC = 1.18) and consistent with polygenicity (see Q–Q plot, Extended Data Fig. 1). We also performed sex-specific GWAS and age-specific GWAS (younger or older than 55 years) because dyslexia prevalence was higher in our younger (5.34% in 20- to 30-year-olds) than older (3.23% in 80- to 90-year-olds) participants. These subsample analyses showed high consistency with the main GWAS (of the full sample). Genetic correlation estimated by linkage disequilibrium (LD) score regression (LDSC) was 0.91 (95% confidence intervals (CI): 0.86–0.96; P = 8.26 × 10−253) in males and females, and 0.97 (95% CI: 0.91–1.02; P = 2.32 × 10−268) between younger and older adults.
https://www.nature.com/articles/s41588-022-01192-y
This is the snapshot of my Dante Labs account with my genome number
I looked at variants with Conditional allele frequency of no more than 10%.
They are based on Grpmax Filtering AF in gnomAD v4
This annotation can be used for filtering variants by allele frequency against a disease-specific threshold that can be set for each disease (e.g. BA1 in the 2015 ACMG/AMP guidelines). In this case the filtering allele frequency (FAF) is the maximum credible genetic ancestry group AF (e.g. the lower bound of the 95% confidence interval (CI)). If the FAF is above the disease-specific threshold, then the observed AC is not compatible with pathogenicity. See http://cardiodb.org/allelefrequencyapp/ and Whiffin et al. 2017 for additional information.
https://gnomad.broadinstitute.org/help/faf
I looked for variants with Combined Annotation Dependent Depletion (CADD) scores of at least 10, and so the variants are predicted to be in the top 10% of the most deleterious.
My focus was mainly on the 5 Prime Untranslated Region, 3 Prime Untranslated Region, and Promoter because they are critical regulatory features in genes. I used VEP to see if the variants involve these regulatory features.
exceptions in regards to CADD scores: 5 Prime Untranslated Region (5'UTR) perturbing variants (Premature Start Codon Gain, Premature Start Codon Loss, Premature Stop Codon Gain, Premature Stop Codon Loss, uORF Frameshift)
In molecular genetics, an untranslated region (or UTR) refers to either of two sections, one on each side of a coding sequence on a strand of mRNA. If it is found on the 5' side, it is called the 5' UTR (or leader sequence), or if it is found on the 3' side, it is called the 3' UTR (or trailer sequence). mRNA is RNA that carries information from DNA to the ribosome, the site of protein synthesis (translation) within a cell. The mRNA is initially transcribed from the corresponding DNA sequence and then translated into protein. However, several regions of the mRNA are usually not translated into protein, including the 5' and 3' UTRs.
Although they are called untranslated regions, and do not form the protein-coding region of the gene, uORFs located within the 5' UTR can be translated into peptides.[1]
The 5' UTR is upstream from the coding sequence. Within the 5' UTR is a sequence that is recognized by the ribosome which allows the ribosome to bind and initiate translation. The mechanism of translation initiation differs in prokaryotes and eukaryotes. The 3' UTR is found immediately following the translation stop codon. The 3' UTR plays a critical role in translation termination as well as post-transcriptional modification.[2]
These often long sequences were once thought to be useless or junk mRNA that has simply accumulated over evolutionary time. However, it is now known that the untranslated region of mRNA is involved in many regulatory aspects of gene expression in eukaryotic organisms. The importance of these non-coding regions is supported by evolutionary reasoning, as natural selection would have otherwise eliminated this unusable RNA.
It is important to distinguish the 5' and 3' UTRs from other non-protein-coding RNA. Within the coding sequence of pre-mRNA, there can be found sections of RNA that will not be included in the protein product. These sections of RNA are called introns. The RNA that results from RNA splicing is a sequence of exons. The reason why introns are not considered untranslated regions is that the introns are spliced out in the process of RNA splicing. The introns are not included in the mature mRNA molecule that will undergo translation and are thus considered non-protein-coding RNA.
https://en.wikipedia.org/wiki/Untranslated_region
Ensembl youtube video playlist Gene Regulation - 8 videos
https://www.youtube.com/playlist?list=PLqB8Yx1tGBMbFtUj_3rYxoasRiMqe3ltO
My Sequencing genomic data
My Dante Labs genomic data
TANC2
Predicted to be involved in dense core granule cytoskeletal transport; regulation of dendritic spine development; and regulation of dendritic spine morphogenesis. Predicted to act upstream of or within in utero embryonic development. Located in dendritic spine. [provided by Alliance of Genome Resources, Apr 2022]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=TANC2
Human phenotypes: Visual hallucinations, Progressive cerebellar ataxia, Broad-based gait, Action tremor, Gaze-evoked nystagmus, Truncal ataxia, Poor eye contact, Dysdiadochokinesis, Dysmetria, Postural instability, Depression, Abnormal social behavior,Impaired social interactions, Anxiety, Stereotypic behavior, Scanning speech, Inappropriate behavior, Impaired smooth pursuit, Spastic gait, Agitation, Gait imbalance, Abnormality of ocular smooth pursuit, Clumsiness, Intention tremor, Specific learning disability, Dysmetric saccades, Hyperacusis
https://maayanlab.cloud/archs4/gene/TANC2
rs150415607 17-63426829-C-T
3'UTR Variant in Enhancer
436 out of 152,348 (0.2862%) Condition allele frequency is 0.4305%
16.7
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA292881839
rs1011977467
17-61503627-TAAC-T GRCh37 (17-63426266-TAAC-T GRCh38) (deletion 3 bases)
3'UTR Variant in Enhancer, CTCF binding site
7 out of 152,120 (0.004602%) Condition allele frequency is 0.007896%
12.6
in Dante Labs genomic data
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA292881552
rs117410905 17-62967674-A-G
Intronic Variant in Promoter
416 out of 152,296 (0.2732%) Condition allele frequency is 0.4129%
16.2
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA293240285
rs150281743 17-62967750-C-A
Intronic Variant in Promoter
434 out of 151,536 (0.2864%) Condition allele frequency is 0.4278%
14.5
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA293240350
CALN1
CALN1 (Calneuron 1) is a Protein Coding gene. Diseases associated with CALN1 include D-Glyceric Aciduria and Deafness, Autosomal Recessive 39. Gene Ontology (GO) annotations related to this gene include calcium ion binding. An important paralog of this gene is CABP7.
This gene encodes a protein with high similarity to the calcium-binding proteins of the calmodulin family. The encoded protein contains two EF-hand domains and potential calcium-binding sites. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2008]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=CALN1
Human phenotypes: Visual hallucinations, Action tremor, Progressive cerebellar ataxia, Broad-based gait, Gaze-evoked nystagmus, Poor eye contact, Dysdiadochokinesis, Impaired social interactions, Abnormal social behavior, Depression, Dysmetria, Anxiety, Scanning speech, Truncal ataxia, Impaired smooth pursuit, Dysmetric saccades, Intention tremor, Postural instability, Abnormality of ocular smooth pursuit, Stereotypic behavior, Spastic gait, Resting tremor, Delusions, Unsteady gait, Abnormality of saccadic eye movements, Gait ataxia, Inappropriate behavior, Agitation, Limb ataxia, Psychosis,Insomnia, Postural tremor, Memory impairment, Diminished motivation, Confusion
https://maayanlab.cloud/archs4/gene/CALN1
rs75361033 7-71787396-G-A
3'UTR Variant
3,521 out of 191,420 (1.839%) Condition allele frequency is 7.238%
11.0
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA160749894
rs76435245 7-72441867-C-T
Intronic Variant
440 out of 152,066 (0.2893%) Condition allele frequency is 0.956%
11.4
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA160841942
rs112952850 7-72135505-T-C
Intronic Variant
2,075 out of 152,330 (1.362%) Condition allele frequency is 4.502%
9.99
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA160795091
GGNBP2
GGNBP2 (Gametogenetin Binding Protein 2) is a Protein Coding gene. Diseases associated with GGNBP2 include Chromosome 17Q12 Deletion Syndrome and Renal Cysts And Diabetes Syndrome. Among its related pathways are 17q12 copy number variation syndrome.
Predicted to be involved in spermatogenesis. Predicted to act upstream of or within several processes, including labyrinthine layer blood vessel development; negative regulation of cell population proliferation; and negative regulation of protein phosphorylation. Predicted to be located in cytoplasmic vesicle. Predicted to be active in cytoplasm and nucleus. [provided by Alliance of Genome Resources, Apr 2022]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=GGNBP2
Human phenotypes: Gaze-evoked nystagmus, Progressive cerebellar ataxia, Broad-based gait, Gait imbalance, Delayed gross motor development
https://maayanlab.cloud/archs4/gene/GGNBP2
rs553160456 17-36544950-G-T
5'UTR Variant in Promoter
1,153 out of 148,858 (0.07746%) Condition allele frequency is 2.554%
17.0
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA290120379
TMEM182
Predicted to be integral component of membrane. [provided by Alliance of Genome Resources, Apr 2022]
Negatively regulates myogenesis and skeletal muscle regeneration via its association with ITGB1 (By similarity). Modulates ITGB1 activation by decreasing ITGB1-LAMB1 interaction and inhibiting ITGB1-mediated intracellular signaling during myogenesis (By similarity). ( TM182_HUMAN,Q6ZP80 )
https://www.genecards.org/cgi-bin/carddisp.pl?gene=TMEM182
Human phenotypes: Progressive cerebellar ataxia, Gaze-evoked nystagmus, Clumsiness, Dysmetric saccades, Broad-based gait, Gait imbalance
https://maayanlab.cloud/archs4/gene/TMEM182
rs1384228186 2-102816139-T-C
3'UTR Variant
2 out of 152,240 (0.001314%) Condition allele frequency is 0.0007990%
15.8
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA754383417
MITF
The protein encoded by this gene is a transcription factor that contains both basic helix-loop-helix and leucine zipper structural features. The encoded protein regulates melanocyte development and is responsible for pigment cell-specific transcription of the melanogenesis enzyme genes. Heterozygous mutations in the this gene cause auditory-pigmentary syndromes, such as Waardenburg syndrome type 2 and Tietz syndrome. [provided by RefSeq, Aug 2017]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=MITF
Human phenotypes: Abnormal auditory evoked potentials, Disinhibition, Abnormality of vision evoked potentials, Horizontal nystagmus
https://maayanlab.cloud/archs4/gene/MITF
rs1004785229 3-69967315-A-G
3'UTR Variant
12 out of 232,640 (0.005158%) Condition allele frequency is 0.01032%
12.4
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA77003736
SGCD
The protein encoded by this gene is one of the four known components of the sarcoglycan complex, which is a subcomplex of the dystrophin-glycoprotein complex (DGC). DGC forms a link between the F-actin cytoskeleton and the extracellular matrix. This protein is expressed most abundantly in skeletal and cardiac muscle. Mutations in this gene have been associated with autosomal recessive limb-girdle muscular dystrophy and dilated cardiomyopathy. Alternatively spliced transcript variants encoding distinct isoforms have been observed for this gene. [provided by RefSeq, Jul 2008]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=SGCD
https://maayanlab.cloud/archs4/gene/SGCD
rs397736317 5-156194495-G-GAA GRch37 (5-156767484-G-GAA GRch38) (Insertion 2 bases)
3'UTR Variant
3,875 out of 72,104 (5.374%) Condition allele frequency is 7.846%
10.9
in Dante Labs genomic data
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA10623720
SEMA3F
SEMA3F (Semaphorin 3F) is a Protein Coding gene. Diseases associated with SEMA3F include Neuroma and Megacolon. Gene Ontology (GO) annotations related to this gene include signaling receptor activity and chemorepellent activity. An important paralog of this gene is SEMA3C.
This gene encodes a member of the semaphorin III family of secreted signaling proteins that are involved in axon guidance during neuronal development. The encoded protein contains an N-terminal Sema domain, an immunoglobulin loop and a C-terminal basic domain. This gene is expressed by the endothelial cells where it was found to act in an autocrine fashion to induce apoptosis, inhibit cell proliferation and survival, and function as an anti-tumorigenic agent. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jan 2016]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=SEMA3F
Human phenotypes: Hyperacusis
https://maayanlab.cloud/archs4/gene/SEMA3F
rs192672815 3-50174560-G-A
Intronic Variant in CTCF binding site
431 out of 152,394 (0.2828%) Condition allele frequency is 0.9139%
15.0
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA74587958
5q34 (chr5:160500001-169000000)
GABRA6
GABRA6 (Gamma-Aminobutyric Acid Type A Receptor Subunit Alpha6) is a Protein Coding gene. Diseases associated with GABRA6 include Anxiety and Intraventricular Meningioma. Among its related pathways are Transmission across Chemical Synapses and GABA B receptor activation. Gene Ontology (GO) annotations related to this gene include chloride channel activity and GABA-A receptor activity. An important paralog of this gene is GABRA4.
GABA is the major inhibitory neurotransmitter in the mammalian brain where it acts at GABA-A receptors, which are ligand-gated chloride channels. Chloride conductance of these channels can be modulated by agents such as benzodiazepines that bind to the GABA-A receptor. At least 16 distinct subunits of GABA-A receptors have been identified. [provided by RefSeq, Jul 2008]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=GABRA6
Human phenotypes: Action tremor, Progressive cerebellar ataxia, Gaze-evoked nystagmus, Visual hallucinations, Dysmetric saccades, Scanning speech, Impaired smooth pursuit, Dysdiadochokinesis, Abnormality of ocular smooth pursuit, Intention tremor, Broad-based gait, Dysmetria, Truncal ataxia, Postural tremor, Horizontal nystagmus, Poor eye contact, Abnormality of saccadic eye movements, Limb ataxia, Impaired social interactions, Abnormal social behavior, Depression, Agitation, Gait ataxia, Anxiety, Postural instability, Confusion, Stereotypic behavior, Psychosis, Spastic gait, Dysarthria, Restlessness
https://maayanlab.cloud/archs4/gene/GABRA6
rs937339709 5-161700233-G-A
Intronic Variant
25 out of 152,342 (0.01641%) Condition allele frequency is 0.09184%
13.6
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA131062740






.png)

.png)



.png)

.png)

.png)

.png)



Comments