My Potential Indicators for High Sensitivity based on 2011 Published 'Contributions of Dopamine-Related Genes and Environmental Factors to Highly Sensitive Personality'
This is a blog post about my potential indicators for high sensitivity. It is based on 2011 Published 'Contributions of Dopamine-Related Genes and Environment Factors to Highly Sensitive Personality'. I am a highly sensitive person, and I have been aware of that for over a couple of decades. Even before I realized that I was a highly sensitive, I've been referred to as sensitive in both positive and negative ways. I am a neurodivergent with Dyslexia, Dyspraxia, ADHD. I also have Ataxia which is a rare neurological issue. Neurodivergents do happen. I got Elaine Aron's books about high sensitivity in people which led me to create the MSN group The Highly Sensitive People back in 1999.
I have done whole genome testing with Dante Labs and Sequencing, and so I checked my genome to check for any possible indicators for high sensitivity.
Contributions of Dopamine-Related Genes and Environmental Factors to Highly Sensitive Personality: A Multi-Step Neuronal System-Level Approach
Abstract
Traditional behavioral genetic studies (e.g., twin, adoption studies) have shown that human personality has moderate to high heritability, but recent molecular behavioral genetic studies have failed to identify quantitative trait loci (QTL) with consistent effects. The current study adopted a multi-step approach (ANOVA followed by multiple regression and permutation) to assess the cumulative effects of multiple QTLs. Using a system-level (dopamine system) genetic approach, we investigated a personality trait deeply rooted in the nervous system (the Highly Sensitive Personality, HSP). 480 healthy Chinese college students were given the HSP scale and genotyped for 98 representative polymorphisms in all major dopamine neurotransmitter genes. In addition, two environment factors (stressful life events and parental warmth) that have been implicated for their contributions to personality development were included to investigate their relative contributions as compared to genetic factors. In Step 1, using ANOVA, we identified 10 polymorphisms that made statistically significant contributions to HSP. In Step 2, these polymorphism's main effects and interactions were assessed using multiple regression. This model accounted for 15% of the variance of HSP (p<0.001). Recent stressful life events accounted for an additional 2% of the variance. Finally, permutation analyses ascertained the probability of obtaining these findings by chance to be very low, p ranging from 0.001 to 0.006. Dividing these loci by the subsystems of dopamine synthesis, degradation/transport, receptor and modulation, we found that the modulation and receptor subsystems made the most significant contribution to HSP. The results of this study demonstrate the utility of a multi-step neuronal system-level approach in assessing genetic contributions to individual differences in human behavior. It can potentially bridge the gap between the high heritability estimates based on traditional behavioral genetics and the lack of reproducible genetic effects observed currently from molecular genetic studies.
Dopamine
16 genes in four subsystems of the dopamine (DA) system:
(1) dopamine synthesis (Tyrosine hydroxylase [TH], Dopa Decarboxylase [DDC]), Dopamine beta-hydroxylase [DBH]);
(2) degradation/transport (COMT, MAOA, MAOB, SLC6A3);
(3) dopamine receptor (DRD1, DRD2, DRD3, DRD4, DRD5);
(4) dopamine modulation (4 Neurotensin genes [NLN, NTS, NTSR1, NTSR2]). These genes represent all major genes involved in these four DA subsystems in humans.
Dopamine synthesis involves converting the amino acid tyrosine (via Tyrosine hydroxylase [TH]) to levodopa (L-DOPA), followed by subsequent decarboxylation (by Dopa Decarboxylase [DDC]) to dopamine. Further conversion by Dopamine beta-hydroxylase (DBH) yields norepinephrine in some cells. For the degradation/transport subsystem, released dopamine is directly broken down at the synapse into inactive metabolites by two enzymes, COMT and MAO (including MAOA and MAOB). The dopamine transporter (SLC6A3), a membrane-spanning protein, pumps the neurotransmitter dopamine into the pre-synaptic neuron for reutilization. For the receptor subsystem, we include
all five genes for dopamine receptors. For the modulation subsystem, we focused on Neurotensin genes, the only well characterized system that has been implicated in the modulation of dopamine signaling.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3135587/
I went with a different approach.
I searched for variants with allele frequencies of no more than 15%. I used Genome Aggregation Database (gnomAD) as reference for the allele frequencies.
My focus was mainly on the 5 Prime Untranslated Region (5'UTR), 3 Prime Untranslated Region (3'UTR), and Promoter because they are critical regulatory features in genes. I used Ensembl's Variant Effect Predictor (VEP) to see if the variants involve these regulatory features.
In molecular genetics, an untranslated region (or UTR) refers to either of two sections, one on each side of a coding sequence on a strand of mRNA. If it is found on the 5' side, it is called the 5' UTR (or leader sequence), or if it is found on the 3' side, it is called the 3' UTR (or trailer sequence). mRNA is RNA that carries information from DNA to the ribosome, the site of protein synthesis (translation) within a cell. The mRNA is initially transcribed from the corresponding DNA sequence and then translated into protein. However, several regions of the mRNA are usually not translated into protein, including the 5' and 3' UTRs.
Although they are called untranslated regions, and do not form the protein-coding region of the gene, uORFs located within the 5' UTR can be translated into peptides.[1]
The 5' UTR is upstream from the coding sequence. Within the 5' UTR is a sequence that is recognized by the ribosome which allows the ribosome to bind and initiate translation. The mechanism of translation initiation differs in prokaryotes and eukaryotes. The 3' UTR is found immediately following the translation stop codon. The 3' UTR plays a critical role in translation termination as well as post-transcriptional modification.[2]
These often long sequences were once thought to be useless or junk mRNA that has simply accumulated over evolutionary time. However, it is now known that the untranslated region of mRNA is involved in many regulatory aspects of gene expression in eukaryotic organisms. The importance of these non-coding regions is supported by evolutionary reasoning, as natural selection would have otherwise eliminated this unusable RNA.
It is important to distinguish the 5' and 3' UTRs from other non-protein-coding RNA. Within the coding sequence of pre-mRNA, there can be found sections of RNA that will not be included in the protein product. These sections of RNA are called introns. The RNA that results from RNA splicing is a sequence of exons. The reason why introns are not considered untranslated regions is that the introns are spliced out in the process of RNA splicing. The introns are not included in the mature mRNA molecule that will undergo translation and are thus considered non-protein-coding RNA.
https://en.wikipedia.org/wiki/Untranslated_region
Ensembl data
Ensembl youtube video playlist Gene Regulation - 8 videos
https://www.youtube.com/playlist?list=PLqB8Yx1tGBMbFtUj_3rYxoasRiMqe3ltO
Variants with an allele frequency of less than 15% in the population
Including only those with at least a UTR, Promoter
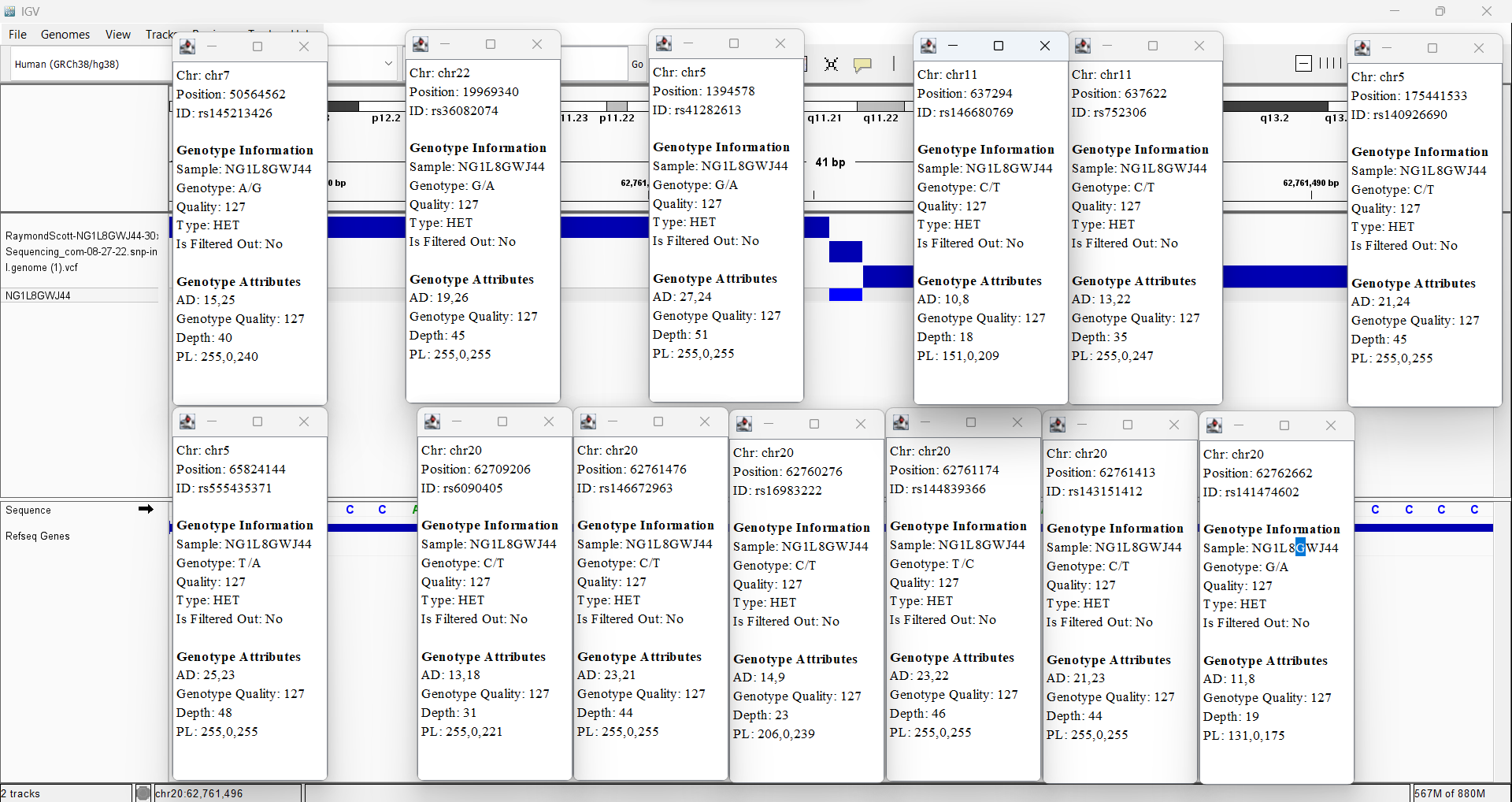
My Sequencing Genomic Data
My Dante Labs Genomic Data
Dopamine Synthesis
DDC
DDC (Dopa Decarboxylase) is a Protein Coding gene. Diseases associated with DDC include Aromatic L-Amino Acid Decarboxylase Deficiency and Autoimmune Polyendocrine Syndrome Type 1. Among its related pathways are Biogenic amine synthesis and superpathway of tryptophan utilization. Gene Ontology (GO) annotations related to this gene include enzyme binding and pyridoxal phosphate binding. An important paralog of this gene is HDC.
The encoded protein catalyzes the decarboxylation of L-3,4-dihydroxyphenylalanine (DOPA) to dopamine, L-5-hydroxytryptophan to serotonin and L-tryptophan to tryptamine. Defects in this gene are the cause of aromatic L-amino-acid decarboxylase deficiency (AADCD). AADCD deficiency is an inborn error in neurotransmitter metabolism that leads to combined serotonin and catecholamine deficiency. Multiple alternatively spliced transcript variants encoding different isoforms have been identified for this gene. [provided by RefSeq, Jun 2011]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=DDC
rs145213426
7-50564562-A-G
Promoter
0.9631%
https://gnomad.broadinstitute.org/variant/7-50564562-A-G?dataset=gnomad_r4
Degradation/Transport
COMT
COMT (Catechol-O-Methyltransferase) is a Protein Coding gene. Diseases associated with COMT include Schizophrenia and Panic Disorder 1. Among its related pathways are Metapathway biotransformation Phase I and II and Neurotransmitter clearance. Gene Ontology (GO) annotations related to this gene include magnesium ion binding and catechol O-methyltransferase activity. An important paralog of this gene is TOMT.
Catechol-O-methyltransferase catalyzes the transfer of a methyl group from S-adenosylmethionine to catecholamines, including the neurotransmitters dopamine, epinephrine, and norepinephrine. This O-methylation results in one of the major degradative pathways of the catecholamine transmitters. In addition to its role in the metabolism of endogenous substances, COMT is important in the metabolism of catechol drugs used in the treatment of hypertension, asthma, and Parkinson disease. COMT is found in two forms in tissues, a soluble form (S-COMT) and a membrane-bound form (MB-COMT). The differences between S-COMT and MB-COMT reside within the N-termini. Several transcript variants are formed through the use of alternative translation initiation sites and promoters. [provided by RefSeq, Sep 2008]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=COMT
rs36082074
22-19969340-G-A
3'UTR, Enhancer, CTCF binding site
2.179%
inherited from Mom
https://gnomad.broadinstitute.org/variant/22-19969340-G-A?dataset=gnomad_r4
SLC6A3
SLC6A3 (Solute Carrier Family 6 Member 3) is a Protein Coding gene. Diseases associated with SLC6A3 include Parkinsonism-Dystonia 1, Infantile-Onset and Tobacco Addiction. Among its related pathways are Transport of inorganic cations/anions and amino acids/oligopeptides and Transmission across Chemical Synapses. Gene Ontology (GO) annotations related to this gene include signaling receptor binding and obsolete protein N-terminus binding. An important paralog of this gene is SLC6A2.
This gene encodes a dopamine transporter which is a member of the sodium- and chloride-dependent neurotransmitter transporter family. The 3' UTR of this gene contains a 40 bp tandem repeat, referred to as a variable number tandem repeat or VNTR, which can be present in 3 to 11 copies. Variation in the number of repeats is associated with idiopathic epilepsy, attention-deficit hyperactivity disorder, dependence on alcohol and cocaine, susceptibility to Parkinson disease and protection against nicotine dependence.[provided by RefSeq, Nov 2009]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=SLC6A3
rs41282613
5-1394578-G-A
3'UTR
0.3551%
inherited from Mom
https://gnomad.broadinstitute.org/variant/5-1394578-G-A?dataset=gnomad_r4
Dopamine Receptor
DRD4
DRD4 (Dopamine Receptor D4) is a Protein Coding gene. Diseases associated with DRD4 include Attention Deficit-Hyperactivity Disorder and Autonomic Nervous System Disease. Among its related pathways are GPCR downstream signalling and Class A/1 (Rhodopsin-like receptors). Gene Ontology (GO) annotations related to this gene include G protein-coupled receptor activity and SH3 domain binding. An important paralog of this gene is DRD3.
This gene encodes the D4 subtype of the dopamine receptor. The D4 subtype is a G-protein coupled receptor which inhibits adenylyl cyclase. It is a target for drugs which treat schizophrenia and Parkinson disease. Mutations in this gene have been associated with various behavioral phenotypes, including autonomic nervous system dysfunction, attention deficit/hyperactivity disorder, and the personality trait of novelty seeking. This gene contains a polymorphic number (2-10 copies) of tandem 48 nt repeats; the sequence shown contains four repeats. [provided by RefSeq, Jul 2008]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=DRD4
rs146680769
11-637294-C-T
5'UTR, Promoter
5.169%
inherited from Mom
https://gnomad.broadinstitute.org/variant/11-637294-C-T?dataset=gnomad_r4
rs752306
11-637622-C-T
Promoter
5.871%
https://gnomad.broadinstitute.org/variant/11-637622-C-T?dataset=gnomad_r4
DRD1
DRD1 (Dopamine Receptor D1) is a Protein Coding gene. Diseases associated with DRD1 include Cerebral Meningioma and Heroin Dependence. Among its related pathways are Class A/1 (Rhodopsin-like receptors) and GPCR downstream signalling. Gene Ontology (GO) annotations related to this gene include G protein-coupled receptor activity and G protein-coupled amine receptor activity. An important paralog of this gene is DRD5.
This gene encodes the D1 subtype of the dopamine receptor. The D1 subtype is the most abundant dopamine receptor in the central nervous system. This G-protein coupled receptor stimulates adenylyl cyclase and activates cyclic AMP-dependent protein kinases. D1 receptors regulate neuronal growth and development, mediate some behavioral responses, and modulate dopamine receptor D2-mediated events. Alternate transcription initiation sites result in two transcript variants of this gene. [provided by RefSeq, Jul 2008]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=DRD1
rs140926690
5-175441533-C-T
3'UTR
0.4598%
https://gnomad.broadinstitute.org/variant/5-175441533-C-T?dataset=gnomad_r4
DRD5
DRD5 (Dopamine Receptor D5) is a Protein Coding gene. Diseases associated with DRD5 include Blepharospasm, Benign Essential and Attention Deficit-Hyperactivity Disorder. Among its related pathways are Class A/1 (Rhodopsin-like receptors) and GPCR downstream signalling. Gene Ontology (GO) annotations related to this gene include G protein-coupled receptor activity and G protein-coupled amine receptor activity. An important paralog of this gene is DRD1.
This gene encodes the D5 subtype of the dopamine receptor. The D5 subtype is a G-protein coupled receptor which stimulates adenylyl cyclase. This receptor is expressed in neurons in the limbic regions of the brain. It has a 10-fold higher affinity for dopamine than the D1 subtype. Pseudogenes related to this gene reside on chromosomes 1 and 2. [provided by RefSeq, Jul 2008]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=DRD5
rs34514924
4-9783730-TA-T (GRChr37: 4-9785354-TA-T)
3'UTR
2.685%
https://gnomad.broadinstitute.org/variant/4-9783730-TA-T?dataset=gnomad_r4
Dopamine Modulation
NLN
NLN (Neurolysin) is a Protein Coding gene. Diseases associated with NLN include Senile Ectropion and Astigmatism. Among its related pathways are Class A/1 (Rhodopsin-like receptors) and GPCR downstream signalling. Gene Ontology (GO) annotations related to this gene include metalloendopeptidase activity and peptide binding. An important paralog of this gene is THOP1.
This gene encodes a member of the metallopeptidase M3 protein family that cleaves neurotensin at the Pro10-Tyr11 bond, leading to the formation of neurotensin(1-10) and neurotensin(11-13). The encoded protein is likely involved in the termination of the neurotensinergic signal in the central nervous system and in the gastrointestinal tract.[provided by RefSeq, Jun 2010]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=NLN
rs555435371
5-65824144-T-A
3'UTR, Enhancer
1.265%
https://gnomad.broadinstitute.org/variant/5-65824144-T-A?dataset=gnomad_r4
NTSR1
NTSR1 (Neurotensin Receptor 1) is a Protein Coding gene. Diseases associated with NTSR1 include Brain Stem Medulloblastoma and Spinal Cord Lipoma. Among its related pathways are Class A/1 (Rhodopsin-like receptors) and GPCR downstream signalling. Gene Ontology (GO) annotations related to this gene include G protein-coupled receptor activity and G protein-coupled neurotensin receptor activity. An important paralog of this gene is NTSR2.
Neurotensin receptor 1 belongs to the large superfamily of G-protein coupled receptors. NTSR1 mediates the multiple functions of neurotensin, such as hypotension, hyperglycemia, hypothermia, antinociception, and regulation of intestinal motility and secretion. [provided by RefSeq, Jul 2008]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=NTSR1
rs6090405
20-62709206-C-T
5'UTR, Promoter, CTCF binding site
9.286%
inherited from Mom
https://gnomad.broadinstitute.org/variant/20-62709206-C-T?dataset=gnomad_r4
rs146672963
20-62761476-C-T
3'UTR, Enhancer
1.316%
https://gnomad.broadinstitute.org/variant/20-62761476-C-T?dataset=gnomad_r4
rs16983222
20-62760276-C-T
3'UTR, Enhancer
0.2181%
https://gnomad.broadinstitute.org/variant/20-62760276-C-T?dataset=gnomad_r4
rs144839366
20-62761174-T-C
3'UTR, Enhancer
1.067%
https://gnomad.broadinstitute.org/variant/20-62761174-T-C?dataset=gnomad_r4
rs143151412
20-62761413-C-T
3'UTR, Enhancer
1.324%
https://gnomad.broadinstitute.org/variant/20-62761413-C-T?dataset=gnomad_r4
rs141474602
20-62762662-G-A
3'UTR
1.075%
https://gnomad.broadinstitute.org/variant/20-62762662-G-A?dataset=gnomad_r4
My Variants in Varient Effect Predictor




.png)




.png)



Comments