My Attention Deficit Hyperactivity Disorder in Connection to Variants in Adhesion G Protein-Coupled Receptor Latrophilin (ADGRL aka LPHN) Genes
I learned that my Attention Deficit Hyperactivity Disorder (ADHD) is in connection to variants in Adhesion G Protein-Coupled Receptor genes. I have three rare Upstream single nucleotide variants in Promoter region in Adhesion G Protein-Coupled Receptor L1 (ADGRL1) aka Latrophilin1 (LPHN1). I have one rare Missense variant, one rare 3 Prime Untranslated Region (3'UTR) single nucleotide variant, two uncommon 5 Prime Untranslated Region (5'UTR) single nucleotide variant in Promoter region and one uncommon 3'UTR single nucleotide variant in Adhesion G Protein-Coupled Receptor L3 (ADGRL3) aka Latrophilin3 (LPHN3). All eight single nucleotide variants have disease-threshold allele frequencies less than the 5% frequency which is the maximum disease-threshold allele frequency that I use for Attention Deficit Hyperactivity Disorder. All have Combined Annotation Dependent Depletion (CADD) scores of at least 10 which means that they are predicted to be in top 10% of most deleterious. Missense variants are moderate-impact variants in coding regions. Upstream variants ,3'UTR variants, and 5'UTR varants are modifier-impact variants in non-coding regions. Promoters and Untranslated Regions have critical regulatory roles in genes.
ADGRL1 (Chromosome 19:14,147,743-14,206,187) and ADGRL3 (Chromosome 4: 61,200,326-62,078,335) are Protein Coding genes that are broadly expressed in the brain. Their molecular functions include transmembrane signaling receptor activity, enabling G protein-coupled receptor activity, enabling protein binding, and enabling carbohydrate binding. Their biological processes include signal transduction, involvement in cell surface receptor signaling pathway, involvement in G protein-coupled receptor signaling pathway, and involvement in adenylate cyclase-activating G protein-coupled receptor signaling pathway, and involvement in cell-cell adhesion via plasma-membrane adhesion molecules. These genes encode a member of the latrophilin subfamily of G-protein coupled receptors (GPCR). Latrophilins may function in both cell adhesion and signal transduction. They were originally discovered as endogenous receptors for α-latrotoxin, a component of black widow spider venom (Latrodectus mactans), but are now known to be involved in brain development and linked to several neuronal and non-neuronal disorders.
According to Malacards, ADGRL1 and ADGRL3 are ranked 3rd and 102nd of the 250 ADHD-related genes listed. The top 15 ADHD-related genes are noted as being elite.
https://www.malacards.org/card/attention_deficit_hyperactivity_disorder_2
https://neurodivergence.blogspot.com/2024/01/the-drd4-dopamine-receptor-d4-gene.html
According to Genecards, some of ADGRL1's Human Phenotypes include Delayed ability to walk, Delayed speech and language development, Attention deficit hyperactivity disorder, Abnormal repetitive mannerisms, Autistic behavior, Delayed ability to sit, Abnormal communication, Language impairment, Atypical behavior, Cognitive impairment, Restricted or repetitive behaviors or interests, Delayed gross motor development, Abnormality of speech or vocalization, Motor delay, Abnormality of the nervous system, Abnormality of mental function, Recurrent maladaptive behavior, Hyperactivity, Short attention span, Disinhibition, Reduced attention regulation, Neurodevelopmental abnormality, Neurodevelopmental delay, Abnormal nervous system physiology, and Abnormal eye physiology. There are no Human Phenotype information for ADGRL3, but I am sure that many of its human phenotypes are same as ADGRL1's human phenotypes.
According Mayaan Lab, some of ADGRL1 and ADGRL3's shared human phenotypes include Progressive cerebellar ataxia, Action tremor, Broad-based gait, Visual hallucinations, Gaze-evoked nystagmus, Truncal ataxia, Dysdiadochokinesis, Dysmetria, Poor eye contact, Anxiety, Inappropriate behavior, Postural instability, Impaired social interactions, Abnormal social behavior, Impaired smooth pursuit, Depression, Scanning speech, Gait imbalance, Abnormality of ocular smooth pursuit, Agitation, Stereotypic behavior, and Spastic gait.
https://maayanlab.cloud/archs4/gene/ADGRL1
https://maayanlab.cloud/archs4/gene/ADGRL3
According to Simon Foundation Autism Research Initiative (SFARI), ADGRL1 is Category 3 (Suggestive Evidence) for Autism.
https://gene.sfari.org/database/human-gene/ADGRL1
The human phenotypes of Adhesion G Protein-Coupled Receptor Latrophilin genes show a significant genetic overlap between ADHD, Dyslexia, Dyspraxia, Autism, Ataxia, and mental illness.
I am a neurodivergent with Dyslexia, Dyspraxia, ADHD, Ataxia, Depression, and Anxiety.
https://neurodivergence.blogspot.com/2024/01/my-neurological-makeup-includes-both.html
Genetics of ADHD
An introduction to the genetics of ADHD by Professor Max Muenke.
ADGRL3 (LPHN3) is discussed and is noted as being related to DRD4 and DRD5.
https://www.youtube.com/watch?v=jQx5ZHn2-fk&t=1260s
https://www.youtube.com/watch?v=HW6zDGwn0Ok&t=34s
ADGRL1
https://pubmed.ncbi.nlm.nih.gov/35907405/
https://www.cell.com/ajhg/pdf/S0002-9297(22)00264-6.pdf
ADGRL3
https://pubmed.ncbi.nlm.nih.gov/32051549/
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5023939/
https://www.nature.com/articles/s41598-022-20343-z
https://www.tandfonline.com/doi/full/10.1080/15622975.2020.1809014
https://www.sciencedirect.com/science/article/abs/pii/S0006322316325768
https://link.springer.com/article/10.1007/s12017-019-08525-x
https://www.biologicalpsychiatryjournal.com/article/S0006-3223(16)32576-8/pdf
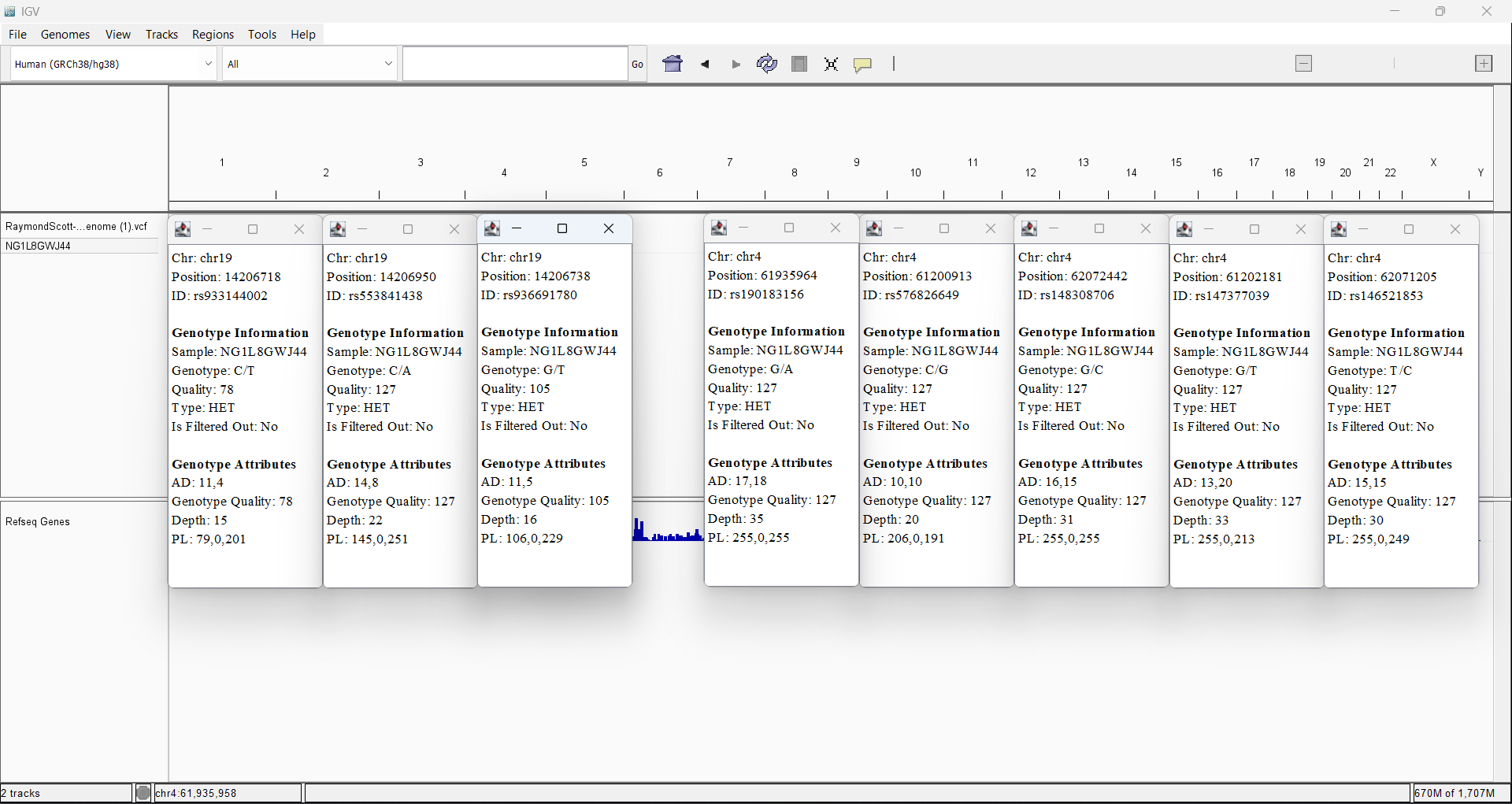
My ADGRL1 variants and ADGRL3 variants from my Sequencing data in Integrative Genome Viewer
ADGRL1 (Chromosome 19:14,147,743-14,206,187)
NCBI Gene Summary for ADGRL1 Gene
ADGRL1 (Adhesion G Protein-Coupled Receptor L1) is a Protein Coding gene. Diseases associated with ADGRL1 include Developmental Delay, Behavioral Abnormalities, And Neuropsychiatric Disorders and Specific Learning Disability. Among its related pathways are GPCRs, class B secretin-like. Gene Ontology (GO) annotations related to this gene include G protein-coupled receptor activity and carbohydrate binding. An important paralog of this gene is ADGRL2.
This gene encodes a member of the latrophilin subfamily of G-protein coupled receptors (GPCR). Latrophilins may function in both cell adhesion and signal transduction. In experiments with non-human species, endogenous proteolytic cleavage within a cysteine-rich GPS (G-protein-coupled-receptor proteolysis site) domain resulted in two subunits (a large extracellular N-terminal cell adhesion subunit and a subunit with substantial similarity to the secretin/calcitonin family of GPCRs) being non-covalently bound at the cell membrane. Latrophilin-1 has been shown to recruit the neurotoxin from black widow spider venom, alpha-latrotoxin, to the synapse plasma membrane. Alternative splicing results in multiple variants encoding distinct isoforms.[provided by RefSeq, Oct 2008]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=ADGRL1
Upstream gene variant in Promoter and Transcription Factor binding site
168 out of 145,788 (0.1152%) Condition allele frequency is 0.3448%
16.6
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA305661423
rs553841438 19-14206950-C-A data unavailable for my mother
Upstream gene variant in Promoter
191 out of 152,074 (0.1256%) Condition allele frequency is 0.3884%
16.5
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA305661588
rs936691780 19-14206738-G-T data unavailable for my mother
Upstream gene variant in Promoter
357 out of 146,710 (0.2433%) Condition allele frequency is 0.4914%
12.5
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA305661457

ADGRL3 (Chromosome 4: 61,200,326-62,078,335)
ADGRL3 (Adhesion G Protein-Coupled Receptor L3) is a Protein Coding gene. Diseases associated with ADGRL3 include Sialuria and Oppositional Defiant Disorder. Among its related pathways are GPCRs, class B secretin-like and GPCRs, other. Gene Ontology (GO) annotations related to this gene include G protein-coupled receptor activity and carbohydrate binding. An important paralog of this gene is ADGRL2.
This gene encodes a member of the latrophilin subfamily of G-protein coupled receptors (GPCR). Latrophilins may function in both cell adhesion and signal transduction. In experiments with non-human species, endogenous proteolytic cleavage within a cysteine-rich GPS (G-protein-coupled-receptor proteolysis site) domain resulted in two subunits (a large extracellular N-terminal cell adhesion subunit and a subunit with substantial similarity to the secretin/calcitonin family of GPCRs) being non-covalently bound at the cell membrane. [provided by RefSeq, Jul 2008]
https://www.genecards.org/cgi-bin/carddisp.pl?gene=ADGRL3
rs190183156 4-61935964-G-A no data available for my mother
Missense variant
279 out of 1,601,982 (0.01742%) Condition allele frequency is 0.3201%
32.0
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA2934690
rs576826649 4-61200913-C-G no data available for my mother
5'UTR Variant in Promoter
1,518 out of 151,884 (0.9994%) Condition allele frequency is 3.349%
14.1
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA98204229
rs148308706 4-62072442-G-C my mother has it too, and so I inherited the variant from her
3'UTR Variant
451 out of 152,532 (0.2957%) Condition allele frequency is 0.9388%
13.0
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA98305708
rs147377039 4-61202181-G-T no data unavailable for my mother
5'UTR Variant in Promoter
1,532 out of 153,544 (0.9978%) Condition allele frequency is 3.368%
10.5
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA98204419
rs146521853 4-62071205-T-C my mother has it too, and so I inherited the variant from her
3'UTR Variant
710 out of 269,412 (0.2635%) Condition allele frequency is 1.331%
10.1
https://reg.clinicalgenome.org/redmine/projects/registry/genboree_registry/by_caid?caid=CA98305586



.png)


Comments